Distribute MATLAB Computations To Python Running on a Cluster with Dask
Part 12 of the Python Is The Ultimate MATLAB Toolbox series.
Introduction
My previous post, Accelerate MATLAB with Python and Numba, showed how compute-bound MATLAB applications can be made to run faster if slow MATLAB operations are rewritten in Python. If the resulting speed improvement isn’t enough, different hardware architectures like GPUs or a cluster of computers may be your only recourse for even higher performance. MATLAB has both of these nailed—if you have the Parallel Computing Toolbox and the necessary hardware.
If you don’t have access to this toolbox
but you do have ssh access to a collection of computers
and you’re willing to
include Python in your MATLAB workflow,
the dask module
can help your MATLAB application tap into those computers' collective CPU power.
- Prerequisites
- Start the dask cluster
- Example 1: sum of prime factors
- Example 2: gigapixel Mandelbrot image
- Example 3: finite element frequency domain response
Prerequisites
Although dask can run on the cores of a single computer, in this article I’ll cover dask’s ability to send computations to, and collect results from a group of remote computers. This raises a few complications: to use dask with a remote computer you’ll need to have an account on it and be able to send commands to it without entering a password. Further, the remote computer should have enough network, file storage, memory, and CPU resources necessary to run your computations. In particular, do you need to share the remote computer with others?
Many organizations with sizeable engineering or research departments
operate compute clusters explicitly to enable large scale parallel
computations. Access and resource sharing are done with job
schedulers such as
LSF,
PBS, or
Slurm.
Dask has
interfaces to these
but here I’ll work with the most simple setups of all, a group
of individual computers on which I have accounts and can
access through ssh.
Here’s the full list of prerequisites:
- compatible (preferrably identical) Python installations including
dask,distributed, andparamikomodules - the same path to the Python executable
- the same account name for the user setting up and using the cluster
- uniform bi-directional, key-based
sshaccess across all computers for the user account - same
sshport - firewall rules open to allow
sshand the dask scheduler and metrics ports (8786 and 8787 by default)
Start the dask cluster
Each computer that is to participate in your computations needs to
have a dask worker process running on it. Additionally, one
of the computers must run the dask scheduler process.
These processes can be started with ssh, Kubernetes, Helm, or
any of the job schedulers (LSF, etc) mentioned above.
Any method other than ssh, however, will likely require help
from a cluster administrator or tech support expert.
I’ll avoid complications and only describe the ssh method.
Note: the method described below of starting a dask cluster with
dask-ssh --hostfileis insecure because anyone with access to the computers and knowledge of the dask port can submit jobs as though they were you. The secure way to set up a dask cluster is with a dask gateway.
Start by creating a text file with each host’s IP address (or hostname, if name resolution works) on a single line. The file might look like this
# file: my_hosts.txt
127.0.0.1
192.168.1.39
192.168.1.40
192.168.1.41
192.168.1.65Hostnames may be repeated. I typically run with a file that defines only localhost entries such as
# file: my_hosts.txt
127.0.0.1
127.0.0.1
127.0.0.1while I’m developing a parallel program. I only submit to remote computers after I’m convinced the code works locally.
Next, pass the file of host names to the dask-ssh command to start
the worker on each node, and also a scheduler:
dask-ssh --hostfile my_hosts.txtYou’ll see a stream of messages resembling
---------------------------------------------------------------
Dask.distributed v2022.7.0
Worker nodes: 3
0: 127.0.0.1
1: 127.0.0.1
2: 127.0.0.1
scheduler node: 127.0.0.1:8786
---------------------------------------------------------------
/usr/local/anaconda3/2021.05/lib/python3.8/site-packages/paramiko/transport.py:219: CryptographyDeprecationWarning: Blowfish has been deprecated
"class": algorithms.Blowfish,
[ scheduler 127.0.0.1:8786 ] : /usr/local/anaconda3/2021.05/bin/python -m distributed.cli.dask_scheduler --port 8786
:
(lines deleted)
:
INFO - Register worker <WorkerState 'tcp://127.0.0.1:37661', status: init, memory: 0, processing: 0>
[ scheduler 127.0.0.1:8786 ] : 2022-11-07 20:58:36,128 - distributed.scheduler - INFO - Starting worker compute stream, tcp://127.0.0.1:37661
[ scheduler 127.0.0.1:8786 ] : 2022-11-07 20:58:36,129 - distributed.core - INFO - Starting established connectionNote line of output with scheduler node: 127.0.0.1:8786.
This is the host and port where the scheduler listens for instructions.
We’ll need this value to create a dask client (described below).
The dask-ssh command will tie up your terminal until the
cluster is shut down.
That can be done by entering ctrl-c in the terminal.
The dask scheduler runs a web server that shows lots of useful information about
the state of your cluster. View it by entering your scheduler node’s
hostname or IP address followed by :8787 as a URL in your browser.
In my case that’s
http://127.0.0.1:8787 or
http://localhost:8787.
The view under the Workers tab looks like this:
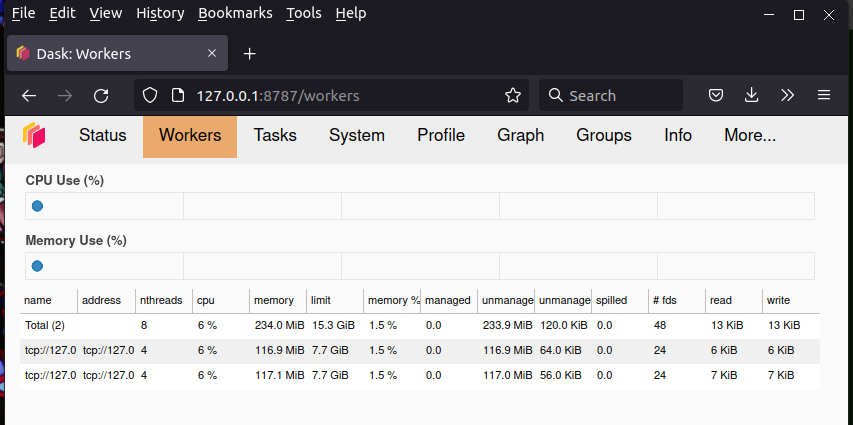
Track your compute nodes on this web while dask is cranking away at one of the following examples, all of which come from the High Performance Computing chapter of my book.
Example 1: sum of prime factors
I’ll start with a simple example to demonstrate the mechanics
of using dask.
The Python and MATLAB programs below compute the sum of
unique prime factors of all numbers in a given range. The hard part,
computing the prime factors themselves, is done with
SymPy’s primefactors() function, comparable to
MATLAB’s factor()1.
Here are sequential implementations:
Python:
|
|
MATLAB:
|
|
Performance looks like this on my laptop:
| Language | Elapsed seconds | Computed sum |
|---|---|---|
| MATLAB 2022b | 546.737 | 5495501355056 |
| Python 3.8.8 | 519.531 | 5495501355056 |
Calls to the compute intensive function my_fn() are independent and
can be called in any order, or even simultaneously.
We’ll do exactly that with dask, using three cores of the local
machine. To spread the load evenly we merely need to offset
the first number in the sequence by the job number, 0 to N-1
for N jobs, then stride through the sequence in steps of N.
For example if we compute prime
factors for numbers between 2 and 20 and want to spread the work
evenly over three jobs, the assignments will be
In : list(range(2,21)) # original set
Out: [2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20]
In : list(range(2+0,21,3)) # for processor 1 (does job 0)
Out: [2, 5, 8, 11, 14, 17, 20]
In : list(range(2+1,21,3)) # for processor 2 (does job 1)
Out: [3, 6, 9, 12, 15, 18]
In : list(range(2+2,21,3)) # for processor 3 (does job 2)
Out: [4, 7, 10, 13, 16, 19]or, equivalently in MATLAB,
>> 2+0:3:20 % for processor 1 (does job 0)
2 5 8 11 14 17 20
>> 2+1:3:20 % for processor 2 (does job 1)
3 6 9 12 15 18
>> 2+2:3:20 % for processor 3 (does job 2)
4 7 10 13 16 19The dask-enabled version of the prime factor summation program on the local computer, splitting up the work to three tasks that are to run simultaneously, looks like this:
|
|
Lines 5, 6 import the dask module and the Client class
Line 16 Creates a Client object that connects to the dask
scheduler on the cluster we set up earlier.
Line 17 Initializes an empty list that will later store “tasks”, basically a unit of work made of a function to call and its input arguments.
Line 19 defines the number of chucks we’ll subdivide the work into. This number should match or exceed the number of dask workers, in other words, the number of remote computers we can run on.
Lines 23-25 populates the task list. Each entry, “job”, is
an invocation of my_fn() with input arguments that stride
between A and B so as to span a unique collection of numbers.
Line 26 tells the scheduler to send the tasks to the workers
to begin performing the computations.
The return value from each task comes back as the list results.
Line 27 is reached when the last worker has finished.
Lines 28-32 fetches the solution for each task and adds it to the global sum variable.
With the scheduler and workers running in the background,
we can now run the
dask-enabled prime factor summation program prime_dask.py.
It runs faster than the sequential version, but not by much:
3 worker processes, 3 dask jobs:
job 0: sum= 2418248946959 T= 327.969
job 1: sum= 659400211060 T= 206.228
job 2: sum= 2417852197037 T= 327.709
total sum=5495501355056
main took 328.852 secPerformance disappoints for two reasons. First, the workload is clearly imbalanced since one job took around 200 seconds, while the other two took more than 320. Evidently, terms of the sequence 3, 6, 9 … can be factored much more rapidly than terms in 2, 5, 8 … and 4, 7, 10 …—who knew? The second reason is less obvious. It is that the sum of individual core times, 328.0 + 206.2 + 327.7 = 861.9 seconds, is 66% higher than the single-core time of 519.5 seconds. Clearly there’s a non-trivial overhead to using dask.
Parallel prime factor summation in MATLAB
Now that we can run our Python program in parallel with dask, we can do the same thing in MATLAB—with a couple of restrictions:
- Tasks submitted to dask must be
Python functions. Therefore our MATLAB main program will
need to send the Python version of the compute function,
my_fn()in this example, to the remote workers. - MATLAB cannot call the
dask.delayed()anddask.compute()functions directly. In particular, the asterisk in the call todask.compute()(see line 26 in the listing above) is not legal MATLAB syntax. We’ll need a small Python bridge module to overcome these limitations.
The first restriction isn’t a big deal for this example; we
merely need to import my_fn() from prime_seq.py or
prime_dask.py. For a more substantial MATLAB application,
though, this could mean translating a lot of MATLAB code
to Python.
Aside: bridge_dask.py
The second restriction is easily resolved with just a few lines of Python:
|
|
This simple module provides wrappers to
dask.delayed() and dask.compute() in a form suitable for MATLAB.
Parallel prime factor summation in MATLAB, continued
We now have the pieces in place to have MATLAB send the prime factor computation and summation work to a cluster of workers. The solution looks like this:
|
|
Line 12 has an important we didn’t need in the pure Python
solution: it uploads our Python module file to the workers.
Keep in mind that the workers do not share memory or file
resources with the main program and therefore need to be given all
items, whether input data or our code files, to function properly.
(Incidentally, the upload_file() function can only be used to
send source files to the workers. A different function, demonstrated
in Example 3, is needed for other files.)
Our bridge module functions are called at lines 18 and 21. Line 26 calls the py2mat.m function to convert the Python solution list into a MATLAB cell array.
MATLAB’s parallel performance matches Python’s—mediocre, not great:
>> prime_dask
Job 0 took 345.014 sec
Job 1 took 219.340 sec
Job 2 took 344.900 sec
A=2 B=10000000, 345.164 sec
S=5495501355056Nonetheless we’ve established the mechanics of sending computations from a MATLAB main program to a collection of Python workers and getting the results back as native MATLAB variables.
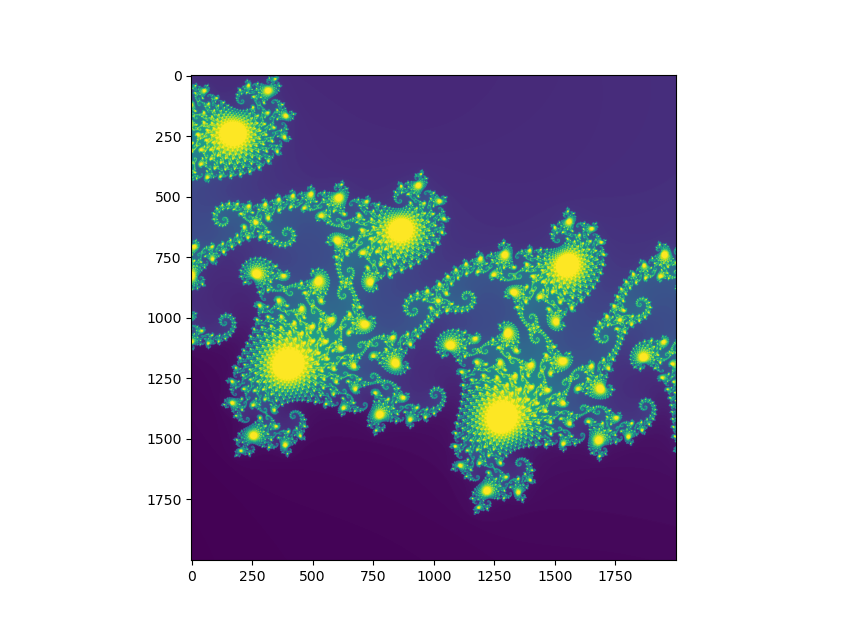
Example 2: gigapixel Mandelbrot image
My second example does a bit more interesting work in a bit
more interesting fashion: I’ll scale up the
Mandelbrot set
example from 5,000 x 5,000 pixels to
35,000 x 35,000—more than 1.2 billion pixels.
This is 49x larger than the 5k x 5k image so we can expect it to take 49x
longer—several minutes instead of several seconds.
This time I’ll farm the work out to actual remote computers,
a collection of
18 virtual machine instances from DigitalOcean2.
Foolishly, perhaps, I
start the dask cluster when I need it using the
insecure dask-ssh --hostname method.
I had to restructure the solution to work on
the memory limited workers.
Instead of simply subdividing my 35,000 x 35,000 frame across
the 18 workers, I had to go an extra step and first break the
frame into smaller sub-frames, spread each sub-frame across the 18 workers,
harvest the results from the sub-frame, then send the next sub-frame out.
The code below uses the term group to refer to a sub-frame:
MATLAB:
|
|
Line 4 imports the Numba-powered Mandelbrot functions from
the file MB_numba_dask.py.
The MB() function in this file differs a bit from the one
in the Python+Numba article.
This MB() figures out which rows of the sub-frame to stride over
then returns the indices of these rows to the caller so that
the caller can reassemble the overall frame more easily.
Line 16 shows the IP address of the localhost. In reality, I use the IP address of the DigitalOcean instance running my dask scheduler. I won’t advertise this address to keep my instances from unnecessary attention.
Line 17 uploads the MB_numba_dask.py module file to the workers.
The loop starting at line 19 iterates over the 16 sub-frames. I further divide each sub-frame across 18 workers so a worker at any one time is only doing math on 35000/(18*16), or roughly 122 rows at a time (each of which has 35000 columns).
Line 28 collects the results for the sub-frame from the 18 workers, then the loop continues to the next sub-frame.
Line 38 and 39 convert the Python numeric solution into native MATLAB variables. These are expensive steps because more than a gigabyte of data has to be copied.
Line 46 displays the resulting image, which doesn’t seem
noteworthy. It is, though. MATLAB on an 8 GB laptop can comfortably
render the gigapixal image whereas matplotlib’s equivalent
command, plt.imshow() crashes.
Here are performance numbers for a typical run:
>> MB_numba_dask
group 0 took 5.658 sec
group 1 took 6.278 sec
group 2 took 4.812 sec
group 3 took 5.495 sec
group 4 took 4.746 sec
group 5 took 5.028 sec
group 6 took 4.982 sec
group 7 took 5.098 sec
group 8 took 4.911 sec
group 9 took 5.036 sec
group 10 took 4.923 sec
group 11 took 4.790 sec
group 12 took 4.946 sec
group 13 took 4.530 sec
group 14 took 4.528 sec
group 15 took 5.192 sec
py2mat conversion took 30.253
111.463 35000In other words, just under 2 minutes to get results when running on 18 computers compared to an estimated 46 minutes for a pure MATLAB solution running on a single computer (56.77 seconds for 5k x 5k * 49 larger problem / 60 sec/min). True, much of the performance comes just from Numba. However the DigitalOcean virtual machines have just one core each and these run much slower than the cores of my laptop. I can’t run MATLAB on the virtual machines so it is difficult to separate know what my parallel efficiency is.
Example 3: finite element frequency domain response
Few real-world problems are as clean to set up and solve as the previous two examples. Realistic tasks typically involve substantial amounts of input data and then create volumes of new data. Figuring out how to split, transfer, then reassemble data efficiently can be as difficult as designing algorithms that can run in parallel. Often, as in this next example, you have to decide whether to transfer data from the main program to the remote workers or have all the workers perform duplicate work to generate the same data. The problem is compounded when using MATLAB with dask because you’ll also need to decide how much of your MATLAB code you’re willing to reimplement in Python for execution on the remote computers.
This example comes from the field of structural dynamics. It solves a system of equations involving stiffness ($K$), damping ($C$), and mass matrices ($M$) from a finite element discretization of a structure at many frequencies $\omega_k$, where $k$ = 1 .. $N$:
$$ K x_k + i \omega_k C x_k - \omega_k^2 M x_k = P(\omega_k) b $$
$N$ is typically in the hundreds but can exceed a thousand if the frequency content of the load spectrum, $P(\omega_k)$, spans a wide range. The vector $b$ stores the static load at each degree of freedom.
If the finite element model is large, each individual solution will take minutes (or worse!). Scale that by $N$ and you’ll be waiting a while for answers, possibly a very long while. The problem is easily divided among processors, however, since each frequency’s solution is independent of the other frequencies.
The first design decision to make is should we 1) compute $K$, $C$, and $M$ in the main program, written in MATLAB, then copy the matrices to the remote computers for solution in Python, or 2) send the model description to the remote computers and have them independently regenerate $K$, $C$, and $M$, then solve them at a subset of frequencies?
The second method involves much less network traffic but it also means translating the MATLAB finite element generation and assembly routines to Python. The first method puts a big load on the network to transfer the large matrices to many remote computers but it also means there’s little extra code to write; Python already has fast linear equation solvers for dense and sparse matrices. I’ll choose the lazy “write less code” option, method 1, in this case.
Chapter 14 of my book
describes the full finite element solver using sparse
matrices implemented in both
MATLAB and Python and includes representative models of a
notional satellite with >600,000 degrees of freedom (DOF).
The elements are merely 2D rods; I made no attempt to emulate
a realistic structure.
Multiple mesh discretizations were created with
triangle.
A 7,600 DOF version
(satellite.2.ele and
satellite.2.node)
looks like this:
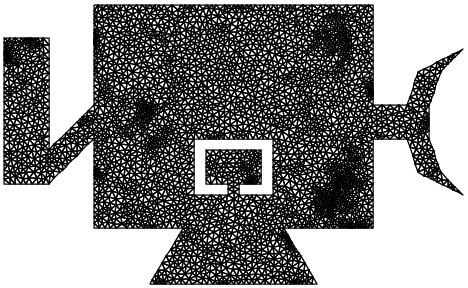
The load represents an explosive bolt in the elbow of the solar array deployment structure on the left. Its time history and spectral content are
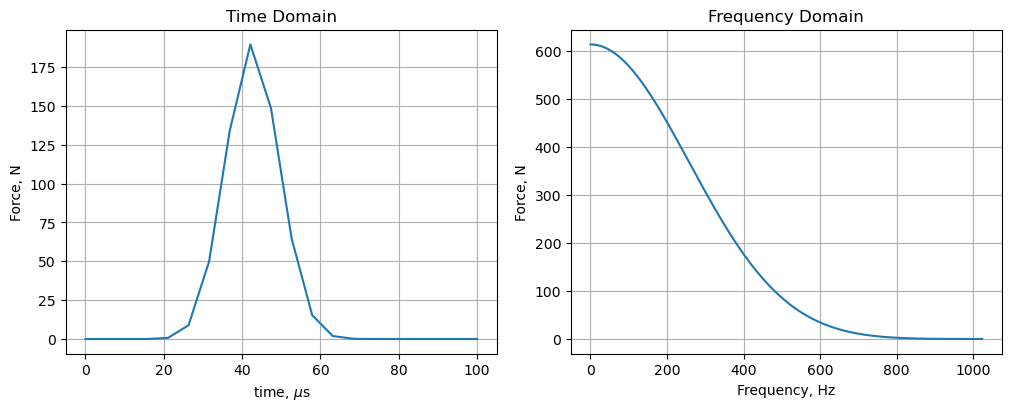
Rather than repeating the details here I’ll describe problems I encountered trying to solve the 600k DOF model in MATLAB + dask across 2,048 frequencies using 100 remote workers of Coiled’s cloud.
Aside: Coiled
Coiled is a company created by dask’s founders. It offers dask as a cloud service. Using this service is much easier path than creating a roll-your-own cluster as I did with my 18 anemic DigitalOcean virtual machines. In addition to providing computationally powerful computers, Coiled gives users 10,000 hours of free CPU each month.
I ran this example on Coiled’s cloud.
Sending large amounts of input data to remote workers
If we have a function that takes two input arguments:
result = my_function(argument_1, argument_2)we can send multiple copies of it to run simultaneously on remote computers with
tasks = []
task = client.submit(my_function, argument_1, argument_2)
tasks.append(task)
task = client.submit(my_function, argument_3, argument_4)
tasks.append(task)
# :
# and so on
# :
results = client.gather(tasks)Dask does many things behind the scenes to make this work. It
- finds an idle worker to run the function
- serializes the function arguments then transmits them over the network to the worker
- deserialized the arguments then instructs the worker to call the function with these arguments
- captures the return values from the function, serializes
them, then transmits them back to the main program where
it deserializes them and adds them to the
resultslist
The second step is problematic when the input arguments are massive and don’t change between function calls. In our case the input matrices $K$, $C$, $M$, $P$, and $b$ are invariant and should only be sent over the network once.
Dask has a function for this situation called scatter()
which sends the given arguments to all remote computers.
It works best when these workers are ready to receive data,
so we’ll use another convenience function, wait_for_workers().
The before and after calls are arranged like this:
Python (regular way, without scatter):
tasks = []
client = Client(cluster)
for i in range(n_jobs):
task = client.submit(solve, K, C, M, b, omega_subset[i])
tasks.append(task)
results = client.gather(tasks)Python (with scatter):
tasks = []
client = Client(cluster)
client.wait_for_workers(n_jobs)
KCMb_dist = client.scatter([K, C, M, b], broadcast=True)
for i in range(n_jobs):
task = client.submit(solve, KCMb_dist, omega_subset[i])
tasks.append(task)
results = client.gather(tasks)MATLAB client disconnects
The next problem I encountered was disconnects between the Python worker processes on the remote computers and the dask client in the MATLAB main program. I still don’t know the cause for this although it may be related to the amount of time the computations took on the remote workers.
The solution I came up with was another Python bridge module that contained all dask objects and function calls. Once all dask activity happened in Python, the MATLAB main program ran successfully.
Source code
The MATLAB code to compute the finite element matrices and run the direct frequency domain problem along with the Python bridge module and equation solver are too lengthy to list here. Instead you can find them at my book’s Github repository in these locations:
| Operation | source file name |
|---|---|
| MATLAB main program | code/dask/run_fr_dask.m |
| Python main program | code/dask/run_fr_dask.py |
| bridge module + solver | code/dask/pysolve.py |
| compute K,M | code/mesh/FE_model.m |
| compute K,M | code/mesh/Node.m |
| compute K,M | code/mesh/Rod_Elem.m |
| compute K,M | code/mesh/load_model.m |
| compute K,M | code/mesh/run_fem.m |
| satellite FE model | satellite |
Performance and cost
The time to solve 2048 frequencies on my 600k DOF model on a single computer in MATLAB is about 8.5 hours. When the MATLAB program distributes the matrices and equation solving step to 100 workers on Coiled’s cloud, the time drops to about 15 minutes, a 33x speed increase.
| Number of Workers | Number of Frequencies | Time [seconds] | Cost [US$] |
|---|---|---|---|
| 3 | 16 | 402.1 | 0.02 |
| 64 | 64 | 472.1 | 0.50 |
| 100 | 100 | 628.0 | 0.88 |
| 100 | 1024 | 743.0 | 0.95 |
| 100 | 2048 | 940.2 | 1.60 |
Concluding remarks
Dask does a lot of work under the hood to make it easy to send work to remote computers. It manages network transmissions of data to and from the workers and balances the load to keep all workers busy. The price for this convenience is a relatively high overhead when tasks are short, that is, less than 10 seconds each. Examples 1 and 2 fall in this relatively wimpy problem class and don’t show dask at its best.
Dask shines on large problems that would otherwise overwhelm a single computer.
Table of Contents | Previous: Accelerate MATLAB with Python and Numba